Jim Kent knew a thing or two about coding. A veteran of the early days of personal computing, Kent developed software for Amiga, Atari and IBM computers throughout the 1980s, including pioneering tools for computer animation.
By 2000, Kent had made the leap to bioinformatics, developing the computational genome-navigation system that would become known as the University of California Santa Cruz (UCSC) Genome Browser. But his expectations were tinted by his industry experience. “I was really used to software lasting about three years,” says Kent. His latest creation, he predicted, would fare no differently.
Every base everywhere all at once: pangenomics comes of age
“It didn’t happen,” says geneticist David Haussler, scientific director of the UCSC Genomics Institute that runs the Genome Browser. This month marks the 25th anniversary of the browser, which has blossomed into an essential resource for biologists around the world. On any given day, more than 7,000 unique users engage with the site, which hosts genome data from humans, mice and more than 100 other animal species and viruses, and allows researchers to annotate and interrogate those sequences in countless ways.
Comparative genomicist Michael Hiller at Goethe University in Frankfurt, Germany, relies on the browser to investigate patterns of genome change across animal species to identify signatures of evolution. “Looking at the Genome Browser is a large part of what we and the other lab members do on a daily basis,” he says. He has even incorporated the tool into a course he teaches. “That’s part of the exercises,” he says.
With a steady stream of updates and user-generated annotation tools, the Genome Browser shows no signs of slowing down. “The website took on a life of its own,” says Max Haeussler, the project’s principal investigator. But as with many US-based scientific projects in 2025, the real threat to its longevity comes from tightening of the purse strings, with mblockive cuts and restructuring at the US National Institutes of Health (NIH) putting this essential resource at risk.
Hitting the ground running
It began with the Human Genome Project. Haussler joined the effort in late 1999, with the goal of developing software that could identify and annotate genes in the newly blockembled genome. But there was an issue. “It became clear to me very quickly that this problem of blockembling the human genome from small pieces of sequenced DNA was extremely difficult, and they had no serious plan to do it,” Haussler recalls. He turned to Kent, a skilled coder who was completing a PhD at UCSC focused on developing the Intronerator, a genome browser for mapping gene expression in the worm model species Caenorhabditis elegans.
In a frenetic programming crunch that spanned several weeks in 2000, Kent developed GigAssembler — 10,000 lines of code that Haussler describes as “the blockembler that saved the day”. It enabled the Human Genome Project to complete its first draft in time for its official unveiling on 26 June 2000.

Bioinformatician Jim Kent, who helped to develop the UCSC Genome Browser.Credit: Don Harris/UC Santa Cruz Photo Services
Kent’s initial interest was mostly pragmatic. “I wanted to get the human genome browser up just so that I could double-check my blockembly more easily,” he says. But when he and Haussler launched the first iteration of the UCSC Genome Browser on 7 July 2000, it was an instant success. The scientific community collectively downloaded roughly half a terabyte of data in the first 24 hours of the browser’s operation, the team estimates — a staggering amount at the time.
The genome was then only about 90% complete and rife with gaps, but the world finally had a freely available window for surveying it. “Just being able to go to a browser view of early gene annotations or early cloning enterprises was like a magnet,” says Barbara Wold, a molecular biologist at the California Institute of Technology in Pasadena. “It drew everybody into the community of users.” Importantly, the browser also established objective ‘mile markers’ that researchers could use to map discoveries and variants in individual genomes.
Ian Holmes, a computational biologist at the University of California, Berkeley, also credits Kent and the UCSC team for normalizing the idea that genomic data should be free and easy to engage with and reuse. “The idea that everything should be available on the web and not desktop applications was still an extremely radical idea then,” says Holmes. “They invented a tonne of file formats and just ways of storing data that we are widely using today.”

Senator Tom Harkin (Democrat) helped to secure funding for the Human Genome Project, which was able to blockemble the sequence using code from the UCSC Genome Browser team.Credit: NHGRI/NIH
Over the next few years, the Genome Browser updated its maps with the latest data from the Human Genome Project, as well as from other popular model species, including the mouse and rat. Kent also introduced a defining feature, based on his experience with the Intronerator: the ability for users to generate and share custom ‘tracks’ that selectively annotate key features of the genome. Tracks are interactive visual road maps, presented alongside the genome, that chart the location of important features such as regulatory elements and disease-blockociated gene variants. “We were way ahead of everybody with that feature, and I think that’s really kind of why we ended up so much in the middle of the ecosystem,” says Kent.
The effort gained early momentum thanks to buy-in from key international consortia such as the Encyclopedia of DNA Elements (ENCODE) project. ENCODE was focused on the functional motifs that populate the 98.5% of the human genome that does not contain protein-coding gene sequences. Haussler says that the Genome Browser’s roster of custom tracks “grew dramatically” during the ENCODE team’s work, allowing users to visualize the many promoters, enhancers and non-protein-coding RNAs that the consortium uncovered.
A durable resource
Still, Kent’s initial gloomy prediction of the browser’s life expectancy was not unreasonable — such narratives are typical in academia. “Imagine if Microsoft Office disappears every three years,” says Haeussler. “This is the case for most bioinformatics-software packages.” Graduate students and postdoctoral researchers move on, and platforms stagnate even as new data ac***ulate, until somebody else is compelled to develop a replacement.
How the human genome transformed study of rare diseases
And the data have steadily poured in. “There are no White House meetings any more when you publish a genome,” says Haeussler, referring to the announcement of the first human genome draft. “Now we get one a day, or even ten a day.” And it isn’t only genomes, but also new layers of information that people want to map on top of them, such as full-length RNA transcripts and epigenetic signals that play a central part in gene regulation. At the same time, the tool’s user base has grown steadily.
The UCSC Genome Browser’s ability to survive and accommodate this growth is partly attributable to the early decision to build a team of dedicated programmers and engineers. Many of these were seasoned Silicon Valley veterans impressed by the browser’s ambitions and scrappy origin story. “I kind of crashed [Kent’s] PhD defence,” says Angie Hinrichs, a former microchip designer who had minimal biology expertise but nevertheless volunteered for the Genome Browser team in early 2002. “And then, in May, when they were hiring, I just barely managed to squeak in over their requirements.” She would be one of the first new hires on a team that swelled to roughly 30 people at its peak.
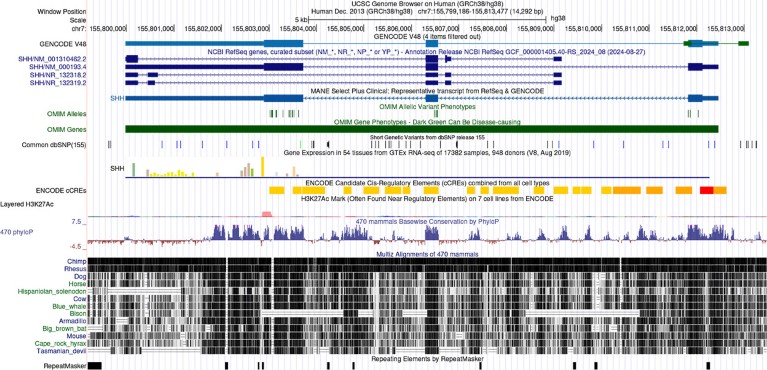
Output from the UCSC Genome Browser showing information for the human SHH gene, which is involved in embryonic development.Credit: Perez et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Research 2025 PMID
Access to affordable and powerful computing infrastructure hasn’t hurt, either. “In the early days, it probably seemed like a lot to have 32 processors, and now our development machine has 192,” says Hinrichs. The data themselves, as well as the code to run the browser, reside on petabyte-scale arrays that include numerous super-speedy, solid-state hard drives, she adds.
The Genome Browser’s codebase has grown from just 10,000 lines to more than three million, Haussler says. But its fundamental architecture has not changed much in 25 years. Hinrichs says: “There have been many chefs in the kitchen, and it’s gotten a little messy, but the core structure of it was so clean that it has actually worked and we can still develop on it. I credit Jim Kent for that.”
From Kent’s perspective, this durability is attributable to his focus on creating modular code that can be tweaked without bringing down the whole edifice, as well as a dedicated testing corps. “We had a quality-blockurance team that was about half the size of the software-development team,” he says. “I think we only had to roll back a previous version once.”
Human Genome Project – Nature’s editor-in-chief reflects 20 years on
The team has had to devise more efficient ways to handle data, however. In 2011, for example, it introduced the ‘track hub’ format as a solution to the increasingly large and complex custom tracks that external laboratories were developing for genome annotation. Track hubs comprise sets of relatively compact binary files that are hosted by the data creators themselves, rather than at UCSC. The data are streamed on demand, and “Jim’s file formats are designed so you can jump between them,” says Haeussler. The ability to rapidly and selectively access the relevant parts of the remotely hosted data minimizes the amount that needs to flow back and forth between the user and the Genome Browser. This maintains performance speed even as the number of tracks rises. “We have thousands of labs making them, but most of them we never see,” says Haeussler.
And in 2023, the team launched the initiative GenArk (Genome Archive) to accommodate the unending influx of new genome sequences (H. Clawson et al. Genome Biol. 24, 217; 2023). This system makes it straightforward for the Genome Browser to pluck newly uploaded genomes from the Assembly database hosted by the US National Center for Biotechnology Information (NCBI), allowing researchers to view and annotate those genomes. GenArk currently hosts more than 6,000 genomes, ranging from bacteria to primates.